Read this article in its entirety in the July/August 2020 digital edition. . .
COVID-19, short for coronavirus disease 2019, is caused by a newly discovered member of the coronavirus family that has been named SARS-CoV-2. This virus was officially recognized as the cause of unexplained and life-threatening respiratory infections observed in December 2019 in China’s Hubei province.
The virus is highly contagious. In just a few months, it spread to nearly every continent in the world and has been declared a global pandemic by the World Health Organization.1 Current estimates, as of the writing of this article, show more than three million cases of COVID-19 have been documented, with over 200,00 of these cases ending fatally—per Microsoft News’ COVID-19 Tracker.
SARS-CoV-2 is a member of the coronavirus family, and this is not the first time that a member of this family has been associated with a life-threatening epidemic. SARS-CoV and MERS-CoV are also viruses from this family that has been very active within the last few decades since their initial identification.1 While these were bad, SARS-CoV-2 is even worse in terms of the total number of infections, fatalities and global reach;1 and the increasing severity of emerging coronaviruses should make us pause to think of the implications of future viral outbreaks.
The coronaviruses were named after their shape was observed under an electron microscope. The viruses are spherical or slightly elliptical and covered with spike-like protrusions, which give them a crown-like appearance (corona is Latin for crown). These spike-like protrusions are in fact glycoproteins that the virus uses to bind to target cells and initiate an infection.2 With respect to SARS-CoV-2, it is thought that a recent mutation (as late as in 2019) in these glycoproteins is the root cause behind the virus’s sudden ability now to infect humans.3
In humans, the SARS-CoV-2 virus targets cells that express the angiotensin-converting enzyme 2 (ACE2) on their plasma membrane.2, 4 Certain areas of the spike-like glycoprotein will attach to membrane-bound ACE2 in the target cell, allowing other areas of the viral glycoprotein to initiate the fusion of the viral particle and the target cell. Infection of potential target cells therefore depends on both the target cell presenting ACE2 on their cell surface and the virus having access to the cell.
Since the virus can survive suspended as an aerosol in water droplets, this makes the most common point of direct interaction between the virus and target cells presenting ACE2 in the respiratory tract, and thus COVID-19 is seen primarily as a respiratory disease. However, ACE2 has a broad expression pattern across many of the organs within the human body.4 It is expressed throughout the cardiovascular system, endocrine system, reproductive system, central nervous system, reproductive system and even the integumentary system. And, while the initial contact between target cells and the virus mainly occurs in the respiratory tract, evidence is emerging that the symptoms of COVID-19 are by no means limited to the respiratory system. Studies are showing that SARS-CoV-2 infections can also cause diarrhea, vomiting, confusion, headache and cardiac injury, demonstrating that once an individual is infected, the virus can spread and infect the different organ systems within the body that express ACE2.5, 6
In addition to the effects noted in the digestive, central nervous and cardiovascular systems, COVID-19 is suspected of infecting the integumentary system. In fact, the current literature is reporting more case studies from individuals diagnosed with COVID-19 showing signs of a wide range of cutaneous lesions.7, 8 As of now, the studies pertaining to the cutaneous effects of COVID-19 are of a general descriptive nature and the exact cell types presumably infected by SARS-CoV-2 in the skin have not been specifically identified—although that may change between now and the publication of this article due to the current worldwide effort to research this virus. However the skin has a fully functional renin-angiotensin system in it, and histological studies have shown that ACE2 expression can be observed in the basal layer of the epidermis and cells of the sebaceous gland,9 suggesting that at the very least, epidermal keratinocytes and sebaceous cells could be at risk of COVID-19 infection.
As the world races to find both vaccines to prevent infections and medications to aid in recovery, there are perhaps some ways in that the skin care industry may assist in limiting the cutaneous effects of SARS-CoV-2 (and by extension, other common skin viral infections). Like most cells in the body, skin cells have an innate immune system with components specifically designed to address viral infections. While these systems can be effective in younger individuals, there is evidence in the literature showing that as humans age, the effectiveness of the skin’s innate immune system begins to decline;10 a phenomenon referred to as immunosenescence.
While the aspects of immunosenescence have been described, there is relatively little research in the current literature addressing the specific mechanisms by which it occurs, or even studies examining how it can be prevented. Given the current worldwide concern for viral infections, this may be an opportune time to correct this deficit in our collective skin care knowledge base of how aging skin loses its ability to respond to viral infection.
Antiviral Mechanisms in Skin
It is well-known the skin is the outermost layer of the body, and in its most basic form, it is a physical barrier between us and external pathogens such as viruses. Yet, despite this protection, there are ways by which viruses can circumvent that physical barrier and gain access to viable cells in the skin; examples include physical trauma to the skin or indirect entry through the respiratory or digestive tracts. Once the virus has access to its target cell, it will bind to it and begin the process of fusing with the cell such that it is able to introduce its nucleic acid cargo into the cytoplasm of the target cell.
Viral genetic information can be stored as either DNA or RNA, and these in turn can be either single or double-stranded. With respect to SARS-CoV-2, its genetic information is stored as a single strand of RNA (+ssRNA), approximately 30 kb in length.1 In humans, our cells have evolved protective mechanisms against these types of viral attacks and within cells, the first line of defense are special receptors that recognize foreign nucleic acids (non-host cell nucleic acids).
With respect to RNA viruses, there are a few prototypical receptor families. Toll Like Receptors (TLR) play a role in innate immune response, and in terms of skin cells, TLR3, TLR7 and TLR9 all react to viral nucleic acids. These receptors are located within either the plasma membrane or cytoplasmic endosomal membranes. TLR3 reacts to double-stranded viral RNA, while TLR7 will react with single-stranded viral RNA.11
While the TLR family of receptors is membrane-bound, there are also receptors within the cytosolic compartment itself. These include receptors such as RIG-1 (retinoic acid-inducible gene 1) and MDA5 (melanoma differentiation associated gene 5). RIG-1 can interact with both single- and double-stranded RNA while MDA5 reacts only with double-stranded RNA.11 These few viral nucleic acid sensors are just a small sampling, and there are many more types within our cells.
Once these viral nucleic acid receptors are activated, they initiate a signaling cascade that travels through many cytoplasmic protein intermediates. In some cases, the signaling cascade involves organelles such as the mitochondria and peroxisomes12 to reach the nucleus and promote one of the main inflammatory signals for viral infection: the formation of interferon.11 The process of the innate response to viral infection up to this point is commonly referred to as the early phase.
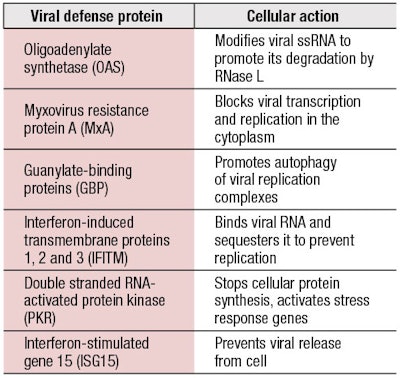
Cells are able to produce many different types of interferon, including alpha, beta, gamma and lambda. While these are all considered interferons, they will work through different types of interferon receptors: Type 1: alpha and beta; Type II: gamma; and Type III: lambda.10 These released interferons can act in both an autocrine and paracrine manner to activate further defenses in both the original infected cell and the neighboring cells, and the different types of interferon receptors will converge in terms of activating a common set of defense proteins commonly referred to as ISGs (interferon-stimulated genes), which includes hundreds of final protein products. The induction of the ISG genes is referred to as the late phase of the innate viral response. The ISG proteins serve an amazing variety of functions with respect to addressing RNA viruses; Table 1 provides just a glimpse of their broad range of capabilities.
As the table suggests, the viral defense proteins are designed to stop the infection at many levels—prevention of the translation of the viral RNA, either by degrading it or by covering it in sequestering proteins; targeted destruction of viral proteins that are produced through autophagy; the total shutdown of cellular protein synthesis; or preventing the release of the newly synthesized viruses. Under normal conditions, these viral defenses can be quite effective but there are times when these simply do not work. As humans have developed many layers of intracellular antiviral defense, viruses have also counter-evolved mechanisms to evade these defenses.13 Evasion methods used by viruses can be incredibly complicated or elegantly simple but research into overcoming these evasion methods normally involves culturing the virus. This is something that most labs in our industry are not set up to do. However, our industry can examine another means by which cellular antiviral defenses may no longer work—by taking a good look at immunosenescence in aging skin.
. . .Read more in the July/August 2020 digital edition. . .
References
- Cascella, M., Rajnik, M., Cuomo, A., et al. (2020 Jan). Features, evaluation and treatment coronavirus (COVID-19) [updated 2020, Apr 6]. in: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing. https://www.ncbi.nlm.nih.gov/books/NBK554776/
- Song, W., Gui, M., Wang, X. and Xiang, Y. (2018, Aug). Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host receptor ACE2. PLOS Pathogens 14(8); e1007236.
- Angeletti, S., Benvenuto, D., Bianchi, Mm, Giovanetti, M., Pascarella, S. and Ciccozzi, M. (2020, Feb 21). COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis. J Med Virology.
- Li, M.Y., Li, L., Zhang, Y. and Wang, X.S. (2020). Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infectious Diseases of Poverty 9(45); https://doi.org/10.1186/s40249-020-00662-x.
- Huang, C., Wang, Y., ... Gu, X., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan China. Lancet; https://doi.org/10.1016/S0140-67636(20)30183-5.
- Chen, N., Zhou, M., ... Wei, Y., et al. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet; https://doi.org/10.1016/S0140-6736(20)30211-7.
- Ahouach, B., Harant, S., ...Bachmeyer, C., et al. (2020, Apr 30). Cutaneous lesions in a patient with COVID-19: Are they related? Brit J Derm; doi: 10.1111/bjd.19168. (e-pub ahead of print)
- Galván Casas, C., Català, A., ... González-Cruz, C. et al. (2020, Apr 29). Classification of the cutaneous manifestations of COVID-19: A rapid prospective nationwide consensus study in Spain with 375 cases. Brit J Derm; doi: 10.1111/bjd.19163. (e-pub ahead of print)
- Grzegrzolka, J., Swiatko, K., ... Podhorska-Okolow, M. (2013). ACE and ACE2 expression in normal and malignant skin lesions. Folia Histochemica et Cytobiologica 51(3) 232-8; doi: 10.5603/FHC.2013.0033.
- Handfield, C., Kwock, J. and Macleod, A.S. (2018, Apr). Innate antiviral immunity in the skin. Trends in Immunol 39(4) 328-340; doi: 10.1016/j.it.2018.02.003.
- Nelemans, T. and Kikkert, M. (2019, Oct 18). Viral innate immune evasion and the pathogenesis of emerging RNA virus infections. Viruses 11(10); doi: 10.3390/v11100961.
- Ferreira, A.R., Marquez, M. and Ribeiro, D. (2019). Peroxisomes and innate immunity: Antiviral response and beyond. Intl J Mol Sci 20 2795; doi:10.3390//ijms20153795.
- Chan, Y.K. and Gack, M.U. (2016, Jun). Viral evasion of intracellular DNA and RNA sensing. Nature Rev Microbio 14(6) 360-73; doi: 10.1038/nrmicro.2016.45 (e-pub 2016, May 13).