Editor’s note: Understanding the complexities of protein folding has perplexed biologists for some 50 years. The way a protein folds into its 3D configuration determines its function—or lack thereof, in the case of some diseases.
Renewed interest (and hope) in protein folding emerged in late 2020 when DeepMind, an AI development firm, announced its AlphaFold system could successfully predict protein folding in 3D. The technology was recognized for this achievement by a group known as the Critical Assessment of protein Structure Prediction (CASP).
“This breakthrough demonstrates the impact AI can have on scientific discovery and its potential to dramatically accelerate progress in some of the most fundamental fields that explain and shape our world,” the firm wrote, in a blog post.
The cosmetic industry picked up on this achievement as well. In fact, some ingredient suppliers have used protein folding as an approach for skin benefits; though its implications may only just be beginning to be realized.
This led us to Karl Linter, Ph.D., an industry leader and pioneer in cosmetic actives and proteins, to whom we posed the question: How could protein folding benefit skin care? Following are his insights.
Theories in Aging
Cosmetic researchers are constantly searching for the newest ingredient and/or mechanism that will allow them to claim anti-X effects for their products; X being age, wrinkles, ptosis, cellulite, hyperpigmentation, hair loss, dark circles and more. A great number of ideas have seen the light in the last four decades—from ingredient families such as AHAs, retinoids, ceramides, peptides and polyphenols, to concepts such as transdermal skin delivery, anti-pollution, the microbiome, epigenetics, mitochondrial disturbance, superfruits, autophagy, glycation, etc.
The number of theories of aging, i.e., causes and symptoms, increases steadily as research in this field advances. But no single theory will turn out to be “the one” because all phenomena are connected; a translation error in the ribosome, a small UV-induced base change in a gene, a loss of mitochondrial efficacy, a pollution related epigenetic change … each and all of these will lead directly or indirectly to further detrimental changes and the well-known symptoms of aging.
Every new discovery of a specific piece of the aging puzzle is scrutinized (and explored) for the possibility of counteracting this process for cosmetic effects. The latest such buzzword is protein folding, found in at least two commercial documentations (described later).
What is this, precisely? And how does it relate to beauty?
What is Protein Folding?
As most readers know, a protein is a long linear chain of amino acids, from a few hundred amino acids to a few thousands. Consider it similar to a strand of pearls or a spaghetti noodle. If the protein is left alone in either a solvent medium of water, in a solution of urea or in cytoplasm, it may look like spaghetti, a random coil or a beautifully complex structure, respectively.
Decades ago, only X-ray crystallography allowed researchers to study the shape of a crystallized protein. Today many other techniques, such as NMR, neutron scattering, fluorescence spectroscopy, superfast algorithms and more yield a “correctly” folded protein structure without the laborious trials of obtaining a good protein crystal for analysis. But what is the “correct” structure? To understand this, it is important to distinguish the four levels of structure in a protein, where each level contributes specifically to the final three-dimensional folded structure (see Figure 1).
Figure 1. The four structures of a protein (By Holger87 - Own work, CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=20899169)
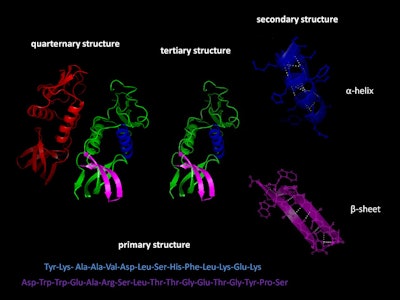 The “primary” structure of a protein is given by its amino acid sequence—the order of the amino acids in the long chain. This sequence, based on the genetic code in DNA, is unique to each protein species and is designated in writing either with the three-letter code, e.g., Tyr-Lys-Ala-Ala-Val … (see Figure 1) or a one-letter code, for long chains; e.g., YKAAV. This sequence does not tell us anything about the shape of the protein—and even less about its function.
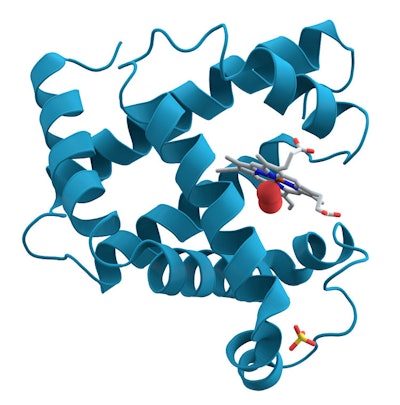
The “primary” structure of a protein is given by its amino acid sequence—the order of the amino acids in the long chain. This sequence, based on the genetic code in DNA, is unique to each protein species and is designated in writing either with the three-letter code, e.g., Tyr-Lys-Ala-Ala-Val … (see Figure 1) or a one-letter code, for long chains; e.g., YKAAV. This sequence does not tell us anything about the shape of the protein—and even less about its function.The first crystallographic analysis of the structure of myoglobin was visualized Max Perutz and John Kendrew, for which they were awarded the Chemistry Nobel Prize in 1962. This elegantly showed that a protein usually does not, in fact, possess a spaghetti shape. The image obtained showed that the protein chain contains eight parts in a tight helical form, and that others appear irregular (see Figure 2).
Figure 2. Myoglobin
 Later, in other proteins, a structure called beta-pleated sheets (see Figure 1) was discovered, followed by beta- and gamma-turns. These details are called the characteristic “secondary” structure of a protein.
Later, in other proteins, a structure called beta-pleated sheets (see Figure 1) was discovered, followed by beta- and gamma-turns. These details are called the characteristic “secondary” structure of a protein.In 1974, the scientists Chou and Fasman of Brandeis University (Massachusetts)1 developed a statistical method based on the sequence of amino acids to calculate the percentage of a-helix and b-pleated sheet structures in proteins. With this, it became obvious to scientists that the side chains of each amino acid and their arrangement along the main protein chain were responsible for the creation of the secondary structure. A replacement of one amino acid in the chain by another, i.e., a transcription error, would often lead to an incorrectly folded protein.
The huge number of protein structures revealed by analysis over the decades has made it clear that the secondary structure is only a tool for the protein to find its next stage: the “tertiary” or 3D structure (see Figure 1). Finally, some special proteins (hemoglobin, for instance) become truly functional only once they are associated in pairs or triplets or quadruplets of themselves in the “quaternary” structure (also see Figure 1).
Why Correct Protein Folding is Important
Christian Anfinsen, Ph.D., of the National Institutes of Health at Bethesda, MD, USA received a Nobel Prize in 1972 for his work on the enzyme ribonuclease and his dogmatic claim that “what a protein does largely depends on its unique 3D structure.” This claim still holds for many proteins but a number of exceptions exist; to quote from Wikipedia:
• “Some proteins need the assistance of chaperone proteins to fold properly.[…]
• However, the chaperones do not appear to affect the final state of the protein; they seem to work primarily by preventing aggregation of several protein molecules prior to the final folded state of the protein. […]
• Many proteins can also undergo aggregation and misfolding […] and some proteins have multiple native structures, and change their fold based on some external factors.”
Once a protein is released from the ribosome where the mRNA has transcribed the DNA sequence into the excreted amino acid chain, how does it “know” in which of the > 10300 (!) or so folded structures at its disposal2 it is supposed to fold? This question has occupied and intrigued scientists for the last 50 years.3
Early experiments, for instance, used a well-characterized enzymatically active protein in solution, then added a sufficient amount of urea to the solution to see the protein lose its activity, its shape and various measurable characteristics; e.g., NMR, IR and CD spectra.4 The removal of the urea could then either bring back the activity of the correctly refolded enzyme, or might lead to a differently shaped but inactive or toxic protein.
Senior, et al.,5 recently claimed, based on major efforts and the help of AI, to have essentially solved the problem of the mechanics of protein folding,6 helping scientists to predict the correct 3D shape of the chain based only on its primary structure. For example, proteins are folded and held together by several forms of molecular interactions that involve thermodynamic stability, hydrophobic and electrostatic Van der Waals interactions, hydrogen and disulfide bonding.
Protein Folding and Skin Aging: Potential Applications
How might all of this biology relate to cosmetics? Keratinocytes and fibroblasts produce many different proteins, constantly. As these cross from the ribosome to and through the endoplasmatic reticulum (ER), they are taken in charge by chaperone proteins, sugar moieties, phosphorylation processes and other cellular tools—all helping the protein to adopt its correct functional shape.
Alas, as skin ages, be it intrinsically or through external aggressions, the reduced efficacy of the ER and decreased number of chaperones leads to increased erroneous protein folding, resulting in increased agglomeration thereof and further reduction in metabolic efficacy in the tissue concerned. Fewer functional proteins may lead to higher rates of translation and transcription errors, and lower production of enzymes and structural proteins such as collagen, elastin, integrins, HA, etc.
As Waller and Maibach7 observe, collagen becomes sparser and is thickened while elastin is degraded, leading to an age-related accumulation of aberrant elastoic material. Generally, age leads to increased [protein] folding […], all interacting in a “feed-forward,” vicious circle.
Silab (France), with its Erisium product (INCI: Oryza Sativa (Rice) Extract), and Mibelle Biochemistry (Switzerland), with its IceAwake (INCI: Succinic Acid (and) Maltodextrin (and) Water (Aqua)) ingredient,8 have explored this avenue for skin care. They refer to academic research on protein processing in the ribosome and endoplasmatic reticulum,9-11 and highlight the fact that aged cells have a tendency to produce incorrectly folded proteins due to reduced amounts of chaperone proteins.
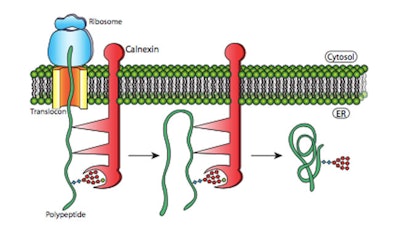
While the Erisium Oryza sativa extract is said to reactivate the UPR (unfolded protein response) pathway, thus reducing ER stress, the IceAwake Iodobacter spp. extract appears to increase chaperone proteins; e.g., BiP, calnexin, calreticulin, endoplasmin—presumably helping proteins to fold correctly and assuring their functionality (see Figure 3).
Figure 3. Protein folding is optimized in the ER.12 Proteins such as calnexin can temporarily bind to nascent polypeptides, preventing them from forming incorrect secondary structures and releasing the protein for folding once the entire polypeptide has been translated.
 Caveat and Conclusion
Caveat and ConclusionProtein folding is a crucial and delicate matter, occurring within the cells, generally tightly controlled by thermodynamics and helped by the aforementioned chaperones. It is presently difficult to measure and visualize this process in living tissue where many ribosomes constantly churn out various proteins as needed.
It is probable that helping the chaperones and the UPR pathway in aged cells in culture with chosen materials, e.g., plants, biotech, synthetic, etc., turns out to be beneficial to the cell and subsequently to the skin. On the other hand, we also must not forget that a number of serious diseases such as transmissible spongiform encephalitis, Creutzfeld-Jacob, scrapie, Alzheimers, etc., are tightly connected with protein folding and its aberrant processes.13
The fact that oxidative stresses are often cited as significantly contributing to these pathologies14 suggests, however, that diverse ingredients such as the cited extracts may exert some preventative and/or curative activities. Further research would shed more light on this new approach.