It is not surprising that skin, being the body’s largest organ and having constant exposure to the environment, is an ideal location for bacteria growth. Such growth includes resident and transient pathogenic bacteria, which are capable of invading the host and causing harm, as well as commensal bacteria, which help to protect the host from pathogens. The skin provides a first line of defense against pathogens based on its mechanical rigidity and low moisture content, production of lysozyme, acidic environment, host defense peptides such as defensins, and as noted, protective commensal microbes.1 Yet, some organisms can and do evade cutaneous host defenses, leading to the next line of protection, which involves the immune system. This leads to the need for preservation and antimicrobial efficacy, with the ultimate goal of ensuring product stability and consumer protection.
In the past, cosmetic formulators used preservatives and biocides for the sole purpose of inhibiting microbes. The Personal Care Products Council, for example, defined a preservative system as any agent added to a product to reduce or prevent microbial growth.2 Antimicrobial protection can be achieved by preserving the products applied to skin, and/or by eliminating harmful microorganisms that already inhabit the skin. However, it is essential to bear in mind the skin’s protective microbiome because the choice of preservative could unintentionally alter the skin’s natural defenses.
Take traditional biocides such as triclosan, for example, which are used in hand sanitizers to disrupt bacterial cell walls. While providing broad-spectrum bactericidal action, they also have nonspecific targets and disturb the skin’s microflora balance, killing both pathogenic and commensal bacteria and leaving the skin defenseless against new destructive microorganisms.3 Triclosan also can cause dangerous antimicrobial resistance to medicines,4 which is a growing threat to health care as a whole. In addition, traditional preservatives such as parabens and formaldehyde donors historically work well at inhibiting microbial growth, but it is not currently known whether they affect or alter the body’s protective microbiota.3
While safety studies and cosmetic research over the past decade have generally succeeded in analyzing the potential toxicity of preservatives and biocides to consumers before they reach the market, only more recently have the effects of those products on skin’s protective commensal microbiota been investigated. In relation, some natural preservatives have been found effective for killing pathogenic bacteria while maintaining commensal microflora on the skin; these are described later. Such elimination of “bad” bacteria and promotion of “good” bacteria is the ideal balance. This article reviews the role of skin microflora in protecting skin, and assesses how the histone deacetylase (HDAC) enzyme is implicated. From this, HDAC expression is used as an indicator to compare the effects of traditional biocides and preservatives with natural antimicrobials on skin’s microbiome.
Understanding Commensal Microflora
Cosmetics and personal care products can support the growth of Gram-negative and Gram-positive bacteria, along with yeasts and mold, but some of these also have nonpathogenic forms that reside as commensal microflora on the skin.5 So the question is, how do antimicrobial agents differentiate between protective and pathogenic microflora, to help maintain the proper balance?
To discuss the safety and effect of biocides and preservatives, it is essential to understand the natural skin microbiome. The diversity of the microbiome begins shortly after birth with the first microbial colonization. With age, the average adult eventually hosts 10 times more microbial cells than human cells,6 which reside in a symbiotic relationship between resident, transient and commensal bacteria. This ecosystem harbors a diverse range of microbial communities that live on physiologically and topographically distinct areas, such as underarms, hands, and feet, as a National Institutes of Health (NIH) microbiome study describes.7
For example, as shown in Figure 1, the primary bacteria located on the human hands include Proteobacteria and Bacteroidetes, both of which are major phylum of Gram-negative bacteria.8 To contrast this, some of the principal microbes located on the feet include Staphylococcaceae and Corynebacterineae; both are very different organisms from those found on different human body sites.
The NIH microbiome study analyzed ribosomal RNA gene sequences obtained from twenty distinct skin sites of healthy humans, and found that physiologically comparable sites harbored similar bacterial communities. These results support the fact that the complexity and stability of a microbial community is dependent on the specific characteristics of the skin site and extrinsic environmental factors with which it comes into contact.7
Some bacteria involved in the microbiome can cause malady; these include resident and transient bacteria, which are capable of invading the protective communities that defend against pathogens. On the other hand, protective or commensal bacteria use competitive inhibition or excretory factors to protect the body from such pathogens. The same NIH study7 not only investigated these differences, but also examined the entire aspect of bacterial symbiosis to explore the microbial interdependencies and cross-talk required to maintain healthy skin.
Recent research has begun to document how such skin commensals interact with one another, pathogenic microbes and human cells. Staphylococcus epidermidis provides a classic example. This commensal Gram-positive bacterium secretes antimicrobial substances that help fight pathogenic invaders. A different Gram-positive bacterium, Propionibacterium acnes, uses the skin’s lipids to produce short-chain fatty acids that can also ward off microbial threats.9
Yasmine Belkaid, of the National Institute of Allergy and Infectious Disease, described the importance of this bacterial cross-talk precisely. “I think anything we can do to restore more balance or more appropriate microbe composition in the skin, as in all the different tissues, is extremely important.”10 These and other skin microbes have an impact on the local molecular environment, and may, as a result, be able to directly alter the behavior of human immune cells.
Histone Deacetylase
An example of how microbes may impact the local molecular environment of skin is provided by a class of enzymes known as histone deacetylases (HDACs). These enzymes help maintain healthy skin by balancing the acetylation activities of histone acetyltransferases on chromatin remodeling, and also playing essential roles in regulating gene transcription.11 Gene transcription leads to protein expression, which plays a crucial part in many physiological pathways such as skin inflammation.
HDAC can be responsive to many different environmental signals and therefore serves as an innovative marker for microflora balance. This is because when the enzyme functions properly, the microbial population of healthy skin remains intact, thus preserving skin’s integrity and natural barrier function. For example, HDAC3, one of the most prominently expressed histone deacetylases in N-TERT human keratinocyte cells, has been shown to have many inflammatory and metabolic roles related to the integrity and function of human organs such as the skin and intestines.12 Once the effects of HDAC on such downstream pathways is better understood, researchers could help consumers to better maintain their skin health by the routine application of products containing antimicrobials.
HDAC and Inflammation
Three publications describing the supporting role HDAC plays in inflammation are of particular interest to the current discussion. First, a study13 from the journal Nature found that HDAC3, while in the presence of commensal bacteria, is a key mediator in maintaining the overall integrity and function of human organs such as the intestines and skin. Major differences were found between the microbial populations in normal mice, compared with those deficient in the HDAC3 enzyme. When this enzyme was reduced, some bacterial species became over-populated, a loss in barrier function was observed, and skin damage and inflammation occurred. In addition, losses of the principle cell type found in epithelium of small intestines were noted.
These results indicate that HDAC plays a key role in the relationship between microbiota and inflammation,13 and that fundamental changes occur in this relationship following the deletion of HDAC3. These changes not only influence the microbiota, but also cell behavior, including downstream gene expression. Without HDAC, a severe loss of barrier function in the large intestine and reduction in colon length also were observed;13 although this experiment focused on internal organs, it is connected with skin care since epithelial barrier functioning applies to both. Further, HDAC is present not only in internal organs but previous research has shown it is expressed in epidermal keratinocyte skin cells (HaCaT).12
A second study,12 from the Journal of Cell Science, investigated HDAC protein levels and total activity dependence on Aryl Hydrocarbon Receptor Nuclear Translocator (ARNT) in N-TERT epidermal keratinocyte cells. Here, the researchers employed an HDAC assay to quantify their findings. In relation, one laba utilized the same methods and assay to compare the microflora integrity of keratinocytes treated with natural vs. synthetic preservatives. This work sought to ascertain whether the expression of HDAC in the epidermis is critical to the maintenance of skin health, keratinocyte differentiation and proper barrier function formation. The study found that ARNT-dependent shifts in HDAC activity could be attributed to significant changes in levels of HDAC proteins present. For example, depletion of ARNT led to an increase in HDAC protein levels and activity, while over-expression of ARNT led to a decrease in HDAC activity.
ARNT is important because it controls both amphiregulin (AREG), the most highly expressed epidermal growth factor receptor (EGFR) ligand in human keratinocytes, and downstream inflammatory pathways, at least in part, by modulating HDAC activity.12 All of these inflammatory and regulatory pathways target advanced stages of epidermal differentiation, and may serve important roles in skin pathologies such as psoriasis and atopic dermatitis. For instance, if a certain biological or chemical stressor is introduced, a change in barrier function is caused by altered immune responses via action on the cytosolic transcription factor aryl hydrocarbon receptor (AhR) and ARNT. The decrease in ARNT then leads to a downstream increase in HDAC level and activity to help restore proper barrier function.
From the research described thus far, it would not be surprising to find that HDAC also plays a key role in the immune and allergic response. However, its role in allergic skin inflammation is not entirely clear. One study from the Journal of Biological Chemistry investigated HDAC and its involvement in the protective inflammatory pathway.14 Results indicated that HDAC, specifically HDAC3, interacted with a specific surface cell protein, FCRI, which contributes to the protective functions of the immune system and regulates the expression of a monocyte chemotactic protein through various gene transcription pathways. These findings suggest that HDAC could serve as a major target for not only developing allergy therapeutics, but also reducing inflammation. This pertains to the cosmetic chemist because ultimately, a major goal among personal care formulators is to eliminate or reduce inflammation as much as possible, especially when using effective levels of preservatives and biocides in cosmetic formulations.
Preservative Mechanisms
Traditional biocides are used to preserve a wide range of products, including cosmetics and personal care as well as foodstuffs and medicines. Examples include disinfectants, antiseptics, pesticides, herbicides, etc. Although these preservatives all serve to protect the consumer, their mechanisms of action differ.3 Activities can range from preserving against multiple varieties of microorganisms, to different strains of one common species. An adapted chart3 from the Journal of Applied Microbiology classifies microbial susceptibilities to biocides, and provides a useful guide for choosing the appropriate biocide for a specific application.
As an example, this chart describes the Gram-positive bacteria Staphylococcus aureus, which has a relatively low resistance to biocides and is therefore usually sensitive to effective use levels of antimicrobial products. On the other hand, highly resistant bacteria such as Gram-negative Providencis species are slightly more resistant and therefore may be harder to kill or inhibit growth.
Another example is triclosan, one of the most effective biocides on the market. It is often used topically as a biocide in antimicrobial products, rather than as a cosmetic preservative. Triclosan is most often associated with hand sanitizers since it has been the main active in them for more than 40 years. Its use in the last 20 years has expanded to soap, cosmetics and toothpaste.15 It is also found in unexpected places; at some retailers, for instance, grocery carts have incorporated it into the handles to prevent bacteria growth.
Although effective, triclosan also has some potentially harmful effects. It works by nonspecifically disrupting bacterial cell walls,4 which inevitably results in the disturbance of the skin’s microflora balance. As the present work will show, triclosan affects HDAC expression in keratinocytes, which denotes a negative impact on commensal microflora and skin barrier function.
Considering these effects, along with continued demand for natural and sustainable ingredients, there are other approaches to consider. Natural antimicrobial peptides are one example. These are typically short chains of less than 50 amino acids, which are synthesized via fermentation for different effects. Those acting in a protective manner secrete chemicals that are cytotoxic to pathogens; others act by a competitive advantage, inhibiting and limiting the growth of pathogens. In both cases, antimicrobial peptides are equipped to kill pathogenic bacteria with proven efficacy. However, the question remained as to whether they affect HDAC activity.
Materials and Methods
Minimum inhibitory concentration (MIC) assays and challenge tests were performed to test the efficacy of a specific antimicrobial peptide (INCI: Leuconostoc/Radish Root Ferment Filtrate). In addition, to compare its effects with synthetic preservatives on skin microflora, an assay using HDAC as a marker was conducted.
MIC assay: MIC assays measure the lowest concentration of an antimicrobial required to inhibit the visible growth of a specific micro-organism after incubation.
Challenge test: Challenge tests evaluate an ingredient’s ability to kill or prevent the growth of microorganisms over time in a finished formula. In this case, two-month, double challenge tests were performed using generic cream formulas. Initially, products were inoculated with micro-organisms and decreases in inoculum were recorded. After the first month, the products were re-inoculated and results were recorded again.
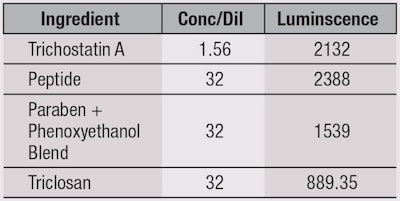
HDAC assay: An HDAC assay was used to screen four ingredients for their effects on HDAC activity as a marker for skin microflora: the test antimicrobial peptide, triclosan, a paraben and phenoxyethanol blend, and Trichostatin A—a known HDAC inhibitor—as a positive control. Serial dilutions of 1:2 were used. This single-reagent-addition, homogenous, luminescent assay is performed via a simultaneous reaction that measures the relative activity of HDAC enzymes from cells.16 The basic chemistry of this assay is depicted in Figure 2.
In brief, HDAC activity deacetylates a luminogenic substrate,16 rendering it sensitive to a specific proteolytic cleavage event that is mediated by the reagent and liberates aminoluciferin. Free aminoluciferin can then be measured using the firefly luciferase reaction to produce a stable, persistent emission of light that can be measured via colorimeter. All three enzymatic events occur in coupled, homogeneous, nearly simultaneous reactions that reach a steady state in 15–45 min.16 Essentially, more HDAC inhibition yields a lower luminescence value, signifying the most damaging antimicrobial as far as the skin microbiome is concerned.
Results
Regarding the MIC, 1-3% of the antimicrobial peptide effectively inhibited the visible growth of the nine microbes tested, as shown in Table 1. Results of the challenge test, shown in Table 2, indicate all microbial growth was reduced by < 99.95%, which is considered successful protection of the generic cream, according to PCPC limits. Also, as seen in Table 3 and Figure 3, the results from the HDAC assay indicate the antimicrobial peptide showed the best HDAC activity, with triclosan performing the worst. The lowest concentration used is depicted because the more concentrate/less dilute the antimicrobial sample, the more toxic it would be to cells in regard to HDAC inhibition. This correlates with the proper dilution factor of tested antimicrobials. It is important to note that these results cannot be compared with untreated media, as no HDAC activity would be present. These findings confirmed the antimicrobial peptide maintained HDAC levels best, and thus would maintain microflora balance and overall skin health better than synthetic compounds such as triclosan and parabens.
Conclusions
In this study, HDAC was evaluated as a novel indicator for skin microflora balance since previous research has shown that when this enzyme is altered or reduced, skin’s commensal microbiota, which protect against unwanted microbes, are less effective. The research presented here proves that some traditional biocides and preservatives, although effective antimicrobials, actually decrease HDAC expression within human skin cells. This could lead to a compromised immune defense system and overall reduced skin health.
However, natural antimicrobials provide a solution to balance effective preservation with maintenance of the skin’s natural microbiome. This promotes the ultimate goal of eliminating pathogenic microbes while maintaining skin’s protective microflora balance, in order to support overall skin health. More research should be performed in this field; however, there is ample promise with the HDAC enzyme as a model for the complex nature of skin and the organisms that live there.
References
- K Chiller et al, Skin microflora and bacterial infections of the skin, J Inves Derm Symp Proceedings 6(3) 170–174 (2001)
- http://webdictionary.personalcarecouncil.org/jsp/Home.jsp (Accessed Mar 27, 2015)
- J-Y Maillard, Bacterial target sites for biocide action, J Applied Microbio Symp Supplement 92 16S-27S (2002)
- SP Yazdankhah et al, Triclosan and antimicrobial resistance in bacteria: An overview, Microb Drug Resist 12(2) 83-90 (2006)
- K Findley et al, Human skin fungal diversity, Nature 498 (7454) 367-370 (2013)
- S Baron, Medical microbiology, U of Tex Medical Branch at Galveston 4, ch 6 (1996)
- E Grice et al, Topographical and temporal diversity of the human skin microbiome, Science 324(5931) 1190-1192 (2009)
- www.genome.gov/dmd/img.cfm?node=Photos/Graphics&id=85320 (Accessed Mar 27, 2015)
- www.the-scientist.com/?articles.view/articleNo/40228/title/Microbes-of-the-Skin/ (Accessed Mar 27, 2015)
- www.the-scientist.com/?articles.view/articleNo/40600/title/The-Body-s-Ecosystem/#skin (Accessed Mar 27, 2015)
- T Akimova et al, Histone/protein deacetylases and T-cell immune responses, Blood J 119(11) 2443-2451 (2012)
- E Robertson et al, ARNT controls the expression of epidermal differentiation genes through HDAC- and EGFR-dependent pathways, J Cell Sci 125 3320-3332 (2012)
- T Alenghat et al, Histone deacetylase 3 coordinates commensal-bacteria-dependent intestinal homeostasis, Nature 504 153-157 (2013)
- K Youngmi et al, histone deacetylase 3 mediates allergic skin inflammation by regulating expression of MCP1 protein, J Biol Chem 287(31) 25844-25859 (2012)
- C Cooney, Triclosan comes under scrutiny, Envt Health Persp 118(6) A242 (2010)
- www.promega.com/~/media/files/resources/protocols/technical%20manuals/101/hdac%20glo%20i%20ii%20assay%20and%20screening%20system%20protocol.pdf (Accessed Mar 27, 2105)